in vitro 3D placenta modelling
mimic the placenta in vitro
in vitro placenta modelling:
|
The overall objective is to develop state-of-the-art models to mimic the placenta in vitro.
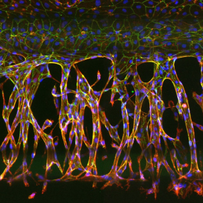
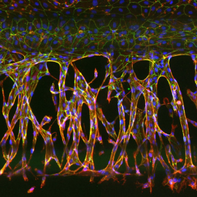
The placenta rapidly develops structurally and functionally throughout gestation and is a unique organ, having blood supply from two organisms. The placenta is made up of maternal and fetal vessels, with numerous highly branched villi lined with trophoblasts (syncytio) facilitating gaseous, nutrient and waste exchange.
Currently, tissue culture using explants from delivered placenta offers the only routine method to study the whole multicellular placenta ex vivo. This restricts our ability to understand the dynamic cellular interaction in normal and diseased placenta, since the explants are from delivered term placenta and are no longer under the same haemodynamic forces.
|
Organ-on-a-chip technology offers functionality not possible with conventional 2D or 3D culture; it is a rapidly advancing technology with the ability to better replicate physiological complexity. MIMETAS OrganoPlates® platform permits advanced organ-on-a-chip technology to be used in any lab setting, providing in vitro modelling of complex multicellular organs. They have successfully developed human perfusable 3D vasculature, CNS, gut and kidney. A placenta-on-a-chip model will be derived from this unique technology to better replicate early-pregnancy events. A combination of stem cell and gene editing technology will drive the development of cell models, encapsulating the complex pathophysiological disease conditions, and implementable in the chip.
Since the 'discovery' of extracellular vesicles (EVs) in 1946 by Erwin Chargraff and Rudolph West interest in those novel cell to cell communicators has risen in almost all fields of biology. EVs have been known to be implicated in diverse physiological and pathological processes and are being considered as a new form of intracellular messaging. There is a significant lack of research in the EV field which focuses on their role during embryonic development and in the pathophysiology of pregnancy complications. During pregnancy EVs cross the placental membrane and establish communication between the mother and the foetus. EVs from amniotic fluid and placental explants will be extracted and characterised and their angiomodulatory signature investigated. By collaboration within the iPLACENTA network the profile of EVs in ex-vivo, in vitro and in vivo systems will be studied. This will allow us to pinpoint their involvement in the development of healthy and diseased pregnancies and development of new methods within the EV field.
Research projects & supervisors
- ESR1 Design and development of placenta-on-a-chip model mimicking normal and diseased placenta characteristics (Dr Henriette Lanz & Dr Chee Ping Ng, Mimetas, Leiden, The Netherlands)
- ESR2 Development of assays to measure translational markers to characterize placental vascular on-a-chip model (Dr Henriette Lanz & Dr Chee Ping Ng, Mimetas, Leiden, The Netherlands)
- ESR3 Identification of placental derived angiomodulatory extracellular vesicles (Dr Benedetta Bussolati, University of Turin, Italy)
- ESR4 Design and generation of in vitro models of diseased placenta to mimic the pathophysiology of preeclampsia and intra-uterine growth restriction (Dr Colin Murdoch, University of Dundee, UK)
integrative placenta modelling
making sense of multi-omics data using systems biology
integrative placenta modelling:
The genetic factors underlying PE and IUGR susceptibility comprise of both maternal and fetal genetic components leading to placental pathophysiology. Genome-wide technologies and genotyping can identify fetal/placental variants. Epigenetic regulation has gained importance in recent years, playing a large part in normal placental development as well as for specific dysregulations observed in pathological situations, where the placenta plays a central role, such as PE and IUGR. Epigenetic regulation of gene expression (defined as heritable and non-associated with DNA mutations) relies upon three major molecular mechanisms: DNA methylation/demethylation, alterations of post-translational marks apposed on the histones, and the load of non-coding RNA (in particular miRNAs). Establishing the full transcriptomic outcome and regulatory mechanisms will enrich the omics datasets in physiological and pathophysiological outcomes and consequently influences the proteome and metabolome. Our aim is full coverage of the ‘omics’ in relation to early and late onset preeclampsia and intrauterine growth restriction.
Research projects & supervisors
- ESR5 Transcriptomic, placental blueprint from human and mouse preeclampsia or growth restriction and circulating miRNAs/lncRNAs (Prof Leon de Windt, Maastricht University, The Netherlands)
- ESR6 Genome-wide investigation in patients with early/late-onset preeclampsia (Dr Daniel Vaiman & Dr Céline Méhats, Institut Cochin/Inserm, Paris, France)
- ESR7 Axis of placental aging in adverse pregnancy outcomes: telomeres and mitochondria (Dr Fergus McCarthy & Dr Cathal McCarthy, University College Cork, Republic of Ireland)
- ESR8 Epigenetic genome-wide investigation in patients with early/late-onset preeclampsia (Dr Daniel Vaiman & Dr Céline Méhats, Institut Cochin/Inserm, Paris, France)
- ESR9 Integrative placenta: A systems biology approach towards phenotype-specific interactomes for placental function (Prof Olaf Wolkenhauer & Dr Shailendra Gupta, University of Rostock, Germany)
imaging placenta
improving pre-clinical and clinical imaging
imaging placenta:
|
|
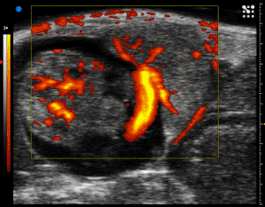
The overall objective is to characterise and validate new methodologies for in vivo placenta modelling and clinical prediction of early/late onset preeclampsia (PE) and intra-uterine growth restriction (IUGR).
Early prediction of pregnancies at risk from PE or IUGR will not only allow stratification of resources: it will provide insight into pathological mechanisms or identification of a window or target for therapeutic intervention.
Animal models have proven to be essential tools furthering our understanding of cardiovascular disease as proof-of-concept, due to the wide availability of different transgenic mouse strains, high level of temporally and cellular genetic control, and the well-documented embryotic development. Yet, animal models investigating PE and IUGR have been restricted by questions of reproducibility and relevance to clinical phenotype. Standardising protocols for in vivo models and characterisation in relation to clinical assessment is essential to prevent duplication of results and discrepancy between scientific findings.
The adaptation of tools is key to providing the ability to assess similar clinical parameters in animal models: Working with Moor Instruments, LSI will be adapted to allow minimally invasive visualisation of organ perfusion such as the placenta to assess perfusion as a surrogate of oxygenation.
|
Moor Instruments develops commercial devices for the measurement of tissue blood flow (Laser Doppler imaging (LDI), fibre optic LD, Laser Speckle contrast imager (LSI)); endothelial function (iontophoresis); and absolute assessment of deep tissue oxygenation (Spatially-resolved NIRS) for a wide range of clinical and research applications.
The ability to evaluate human placental structure and function in real time, by using new advances in ultrasound or magnetic resonance imaging (MRI) in early gestation, will help determine the clinical trajectory of a patient toward the development of placental disorders like PE and IUGR. This has the potential to allow physicians to intervene early and possibly effect/limit the severity of these diseases. All physiological data obtained will be layered onto omics data, generating an advance disease platform for interrogation.
Research projects & supervisors
- ESR10 Development of endoscopic laser speckle imaging probe and miniaturise iontophoresis for assessment of placenta perfusion and endothelial function in murine pregnancy models (Dr Colin Murdoch, University of Dundee, UK & Dr Rodney Gush, Moor Instruments, UK)
- ESR11 Enhancing imaging of the placenta in murine models of preeclampsia and standardisation of preeclampsia models (Prof Asif Ahmed & Dr Keqing Wang, Aston University, Birmingham, UK)
- ESR12 Consequences of intra-uterine growth restriction in feto-maternal hemodynamics and brain development in the rabbit model (Prof Jan Deprest, Katholieke Universiteit Leuven, Belgium)
- ESR13 Application of novel ultrasound technology for improving diagnostic capability of complicated pregnancies (Prof Asma Khalil & Prof Basky Thilaganathan, St George's, University of London, UK)
- ESR14 Development of epigenetic markers of abnormal fetal intrauterine neurodevelopment, correlation with fetal doppler ultrasound (Dr Jose Morales Rosello, Health Research Institute Hospital La Fe (HULAFE), Valencia, Spain
- ESR15 Development of novel ultrasound tools for maternal and fetal cardiovascular assessment (Prof Basky Thilaganathan & Prof Asma Khalil, St George's, University of London, UK)

This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 765274
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 765274